| Editor by Xueyue
| Xi
T cell exhaustion is a state of poor response to tumor antigens, low proliferation and persistence of cells in the body, and induces the expression of inhibitory surface receptors. Initially discovered in the setting of chronic viral infection, T cell exhaustion has subsequently been found to occur in a variety of diseases, including tumors and the autoimmune disease . T cell exhaustion is a major obstacle for checkpoint blockade and CAR-T cell therapies . Manipulating this process improves T cell responses in anti-tumor immunity. Recent studies demonstrate that T cell exhaustion undergoes extensive remodeling of the transcriptional and epigenomes. This unique epigenetic state is primarily driven by chronic antigen and TCR signaling and results in a stable cellular phenotype that is not altered by anti-PD-1 treatment.
On June 23, 2022, Ansuman T. Satpathy team from Stanford University published a paper titled Genome-wide CRISPR screens of T cell exhaustion identify chromatin remodeling factors that limit T cell persistence article. Author constructed a CRISPR screen and found that genes related to epigenetic chromosome remodeling and nucleosome assembly in exhausted T cells are key genes that regulate the exhaustion state.

The authors first developed a genome-wide CRISPR screening assay for depletion in T cells. The authors used anti-CD3 antibodies for 8 days to induce chronic TCR signaling in a manner independent of the antigen . This method can obtain enough cells to cover the entire genome for CRISPR screening. After 8 days of culture, the authors found that PD-1 and TIM-3 were progressively up-regulated. Compared with cells without further TCR stimulation after initial activation, chronically stimulated T cells exhibit growth defects, disrupted secretion of IFNg and TNFa, and defects in tumor killing ability in vivo and in vitro. The authors used ATAC-seq to verify that induced in vitro exhausted T cells displayed similar hallmark functional and genomic features as in vivo exhausted T cells.
Next, the author conducted CRISPR screening on exhausted T cells induced in vitro, and conducted GO analysis on the top 100 exhaustion positive regulators, and found that the TCR signaling pathway was enriched. In addition to GO enrichment related to TCR, other enriched pathways are related to epigenetic , including chromosome remodeling and nucleosome disassembly. The authors also used Cytoscape to visualize the highly enriched gene and depleted gene networks, confirming their highly interconnected relationship networks. Analysis of single-cell transcriptome data from chronic viral infections showed that 98 of the top 100 genes screened were expressed in virus-infected exhausted T cells. The authors selected the BAF and INO80 complexes in the screen as a direction for further research.
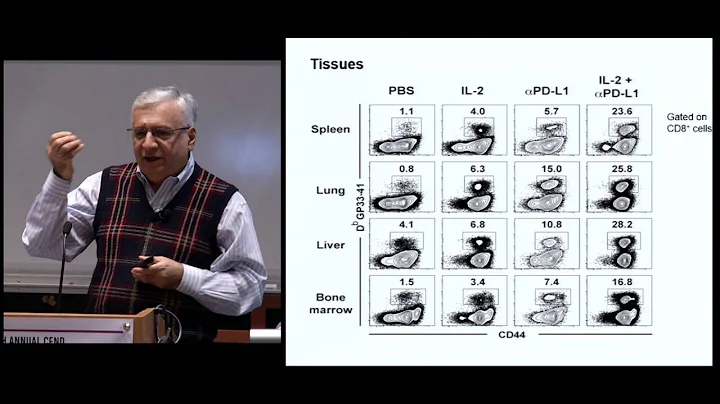
During the screening, the authors observed that Arid1a, Smarcc1 and Smarcd2 are the three most important components of the cBAF complex. After the authors knocked out Arid1a, blocking the cBAF complex could improve T cell persistence and anti-tumor immune efficacy in vivo. This phenomenon has also been verified by the authors in human exhausted T cells. Knockdown of cBAF subunits including Arid1a improves T cell function by limiting the accessibility of AP-1 transcription factors to chromatin, thereby inhibiting the acquisition of the exhaustion-associated chromatin state. Further analysis revealed that cBAF and INO80 chromatin remodeling complexes have distinct roles in T cell exhaustion. cBAF mainly regulates effector and depletion-related genes, while INO80 mainly regulates metabolism. This also suggests that collaboratively targeting multiple pathways can improve T cell function.

This study provides a map of the gene that regulates T cell exhaustion. Using a genome-wide screen, we found that enrichment of chromatin-associated epigenetic factors and nucleosome remodeling were closely associated with exhausted T cells. The work of this article provides a new way to seek to improve T cell function.
Original link:
https://doi.org/10.1016/j.ccell.2022.06.001
Plate maker: 11
Reprinting instructions
[Original article] B ioArt original articles are welcome to be forwarded and shared by individuals. Reprinting without permission is prohibited. The copyright of all published works is owned by BioArt. BioArt reserves all legal rights and any violators will be prosecuted.










![[Source: Shunyi Ecological Environment] Ubiquitous plastic products have facilitated people's lives, but the resulting plastic waste has caused a lot of pollution to the ecological environment... - DayDayNews](https://cdn.daydaynews.cc/wp-content/themes/begin/img/loading.gif)